Getting Started with Pandas
Pandas contains data structures and data manipulation tools designed to make data cleaning and analysis fast and easy in Python.
While pandas adopts many coding idioms from NumPy, the biggest difference is that pandas is designed for working with tabular or heterogeneous data. NumPy, by contract, is best suited for working with homogeneous numerical array data.
1
2
3
| import pandas as pd
from pandas import Series, DataFrame
import numpy as np
|
Introduction to Pandas Data Structures
Series - is a one dimensional array-like object containing a sequence of values and an associated array of data labels, called its index.
1
2
3
4
5
6
7
8
9
10
| obj = pd.Series([4,7,-5,3])
obj
Output:
0 4
1 7
2 -5
3 3
dtype: int64
|
The string representation of a Series displayed interactively shows the index on the left and the values on the right. Since, we did not specify an index for the data, a default one consisting of the integers 0 through N-1 (where N is the length of the data) is created.
We can get the array representation and index objects of the Sereis via its ‘values’ and ‘index’ attributes respectively.
1
2
3
4
5
| obj.values
Output: array([ 4, 7, -5, 3], dtype=int64)
obj.index
Output: RangeIndex(start=0, stop=4, step=1)
|
Often it will be desirable to create a Series with an index identifying each data point
1
2
3
4
5
6
7
8
9
10
11
| obj2 = pd.Series([4,7,-5,3], index=['d','b','a','c'])
obj2
Output:
d 4
b 7
a -5
c 3
dtype: int64
obj2.index
Output:Index(['d', 'b', 'a', 'c'], dtype='object')
|
We can use labels in the index when selecting single values or set of values.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
| obj2['a']
Output: -5
obj2['d']
Output: 4
obj2[['c', 'a', 'd']]
Output:
c 3
a -5
d 4
dtype: int64
obj2[obj2 > 0]
Output:
d 4
b 7
c 3
dtype: int64
obj2 * 2
Output:
d 8
b 14
a -10
c 6
dtype: int64
np.exp(obj2)
Output:
d 54.598150
b 1096.633158
a 0.006738
c 20.085537
dtype: float64
|
Note: Series can also be thought of as a fixed-length, ordered dict, as it is a mapping of index values to data values</p>
1
2
3
4
5
| 'b' in obj2
Output: True
'e' in obj2
Output: False
|
Converting a Python dict obj to a Series obj in pandas.
1
2
3
4
5
6
7
8
9
10
11
| sdata = {'Ohio': 35000, 'Texas': 71000, 'Oregon': 16000, 'Utah' : 5000}
obj3 = pd.Series(sdata)
obj3
Output:
Ohio 35000
Texas 71000
Oregon 16000
Utah 5000
dtype: int64
|
When we pass only a dict, the index in the resulting Series will have the dict’s keys in sorted order. We can override this by passing the dict keys in the order we want them to appear in the resulting Series.
1
2
3
4
5
| type(sdata)
Output: dict
type(obj3)
Output: pandas.core.series.Series
|
1
2
3
4
5
6
7
8
9
| states = ['California', 'Ohio', 'Oregon', 'Texas']
obj4 = pd.Series(sdata, index = states)
obj4
Output:
California NaN
Ohio 35000.0
Oregon 16000.0
Texas 71000.0
dtype: float64
|
Here, 3 values found in sdata were placed in the appropriate locations, but since no value for ‘California’ was found, it appears as NaN(not a number), which is considered in pandas to mark missing or NA values.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
| pd.isnull(obj4)
Output:
California True
Ohio False
Oregon False
Texas False
dtype: bool
pd.notnull(obj4)
California False
Ohio True
Oregon True
Texas True
dtype: bool
|
The ‘isnull’ and ‘notnull’ functions in pandas should be used to detect missing data.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
| obj4.isnull()
Output:
California True
Ohio False
Oregon False
Texas False
dtype: bool
obj3
Output:
Ohio 35000
Texas 71000
Oregon 16000
Utah 5000
dtype: int64
obj4
Output:
California NaN
Ohio 35000.0
Oregon 16000.0
Texas 71000.0
dtype: float64
obj3 + obj4
California NaN
Ohio 70000.0
Oregon 32000.0
Texas 142000.0
Utah NaN
dtype: float64
|
Both the Series object itself and its index have a name attribute, which integrates with other key areas of pandas funcitonality:
1
2
3
4
5
6
7
8
9
10
11
12
| obj4.name = 'population'
obj4.index.name = 'state'
obj4
Output:
state
California NaN
Ohio 35000.0
Oregon 16000.0
Texas 71000.0
Name: population, dtype: float64
|
A series index can be altered in-place by assignment:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
| obj
Output:
0 4
1 7
2 -5
3 3
dtype: int64
obj.index = ['Bob', 'Seteve' ,'Jeff', 'Ryan']
obj
Output:
Bob 4
Seteve 7
Jeff -5
Ryan 3
dtype: int64
|
DataFrame: A DataFrame represents a rectangular table of data and condains an ordered collection of columns, each of which can be a different value type (numeric, string, boolean, etc.). The DataFrame has both a row and column index; it can be thought of as a dict of Series all sharing the same index.
Note: While a DataFrame is physically two-dimensional, we can use it to represent higher dimensional data in a tabular formati using heirarchial indexing.
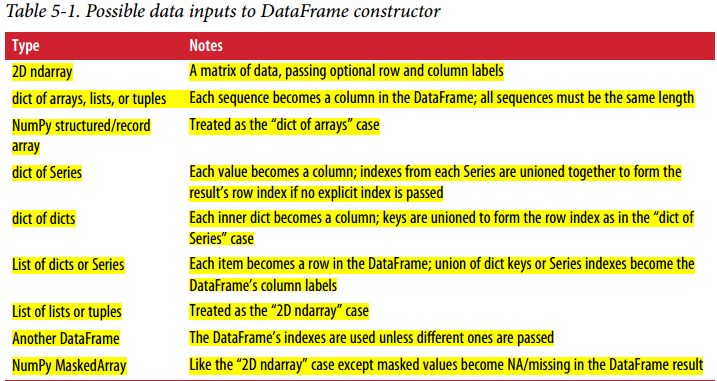
There are many ways to create a DataFrame, though one of the most common is from a dict of equal-length lists or NumPy array:
1
2
3
4
5
6
| data = {'state': ['Ohio', 'Ohio', 'Ohio', 'Nevada', 'Nevada', 'Nevada'],
'year': [2000,2001,2002,2001,2002,2003],
'pop': [1.5,1.7,3.6,2.4,2.9,3.2]}
frame = pd.DataFrame(data)
frame
|
Output:
| | pop | state | year |
| 0 | 1.5 | Ohio | 2000 |
| 1 | 1.7 | Ohio | 2001 |
| 2 | 3.6 | Ohio | 2002 |
| 3 | 2.4 | Nevada | 2001 |
| 4 | 2.9 | Nevada | 2002 |
| 5 | 3.2 | Nevada | 2003 |
For Large DataFrames, the head method selects only the first five rows<
Output:
| | pop | state | year |
| 0 | 1.5 | Ohio | 2000 |
| 1 | 1.7 | Ohio | 2001 |
| 2 | 3.6 | Ohio | 2002 |
| 3 | 2.4 | Nevada | 2001 |
| 4 | 2.9 | Nevada | 2002 |
If we specify a sequence of columns, the DataFrame’s columns will be arranged in that order:
1
| pd.DataFrame(data,columns=['year', 'state', 'pop'])
|
Output:
| | year | state | pop |
| 0 | 2000 | Ohio | 1.5 |
| 1 | 2001 | Ohio | 1.7 |
| 2 | 2002 | Ohio | 3.6 |
| 3 | 2001 | Nevada | 2.4 |
| 4 | 2002 | Nevada | 2.9 |
| 5 | 2003 | Nevada | 3.2 |
If passed a column that isn’t contained in the dict, it will appear with the missing values in the result:
1
2
3
| frame2 = pd.DataFrame(data, columns = ['year', 'state', 'pop', 'debt'],
index=['one', 'two', 'three', 'four', 'five', 'six'])
frame2
|
Output:
| | year | state | pop | debt |
| one | 2000 | Ohio | 1.5 | NaN |
| two | 2001 | Ohio | 1.7 | NaN |
| three | 2002 | Ohio | 3.6 | NaN |
| four | 2001 | Nevada | 2.4 | NaN |
| five | 2002 | Nevada | 2.9 | NaN |
| six | 2003 | Nevada | 3.2 | NaN |
1
2
| frame2.columns
Output: Index(['year', 'state', 'pop', 'debt'], dtype='object')
|
We can retrive a column in a DataFrame as a Series either by dict-like notation or by attribute:
1
2
3
4
5
6
7
8
9
10
| frame2['state']
Output:
one Ohio
two Ohio
three Ohio
four Nevada
five Nevada
six Nevada
Name: state, dtype: object
|
1
2
3
4
5
6
7
8
9
| frame2.year
Output:
one 2000
two 2001
three 2002
four 2001
five 2002
six 2003
Name: year, dtype: int64
|
Rows can also be retrieved by position or name with the special loc attribute .
Output:
| | year | state | pop | debt |
| one | 2000 | Ohio | 1.5 | NaN |
| two | 2001 | Ohio | 1.7 | NaN |
| three | 2002 | Ohio | 3.6 | NaN |
| four | 2001 | Nevada | 2.4 | NaN |
| five | 2002 | Nevada | 2.9 | NaN |
| six | 2003 | Nevada | 3.2 | NaN |
1
2
3
4
5
6
7
| frame2.loc['three']
Output:
year 2002
state Ohio
pop 3.6
debt NaN
Name: three, dtype: object
|
Columns can be modified by assignment. For example, the empty debt column could be assigned a scalar value or an array of values:
1
2
3
|
frame2['debt'] = 16.5
frame2
|
Output:
| | year | state | pop | debt |
| one | 2000 | Ohio | 1.5 | 16.5 |
| two | 2001 | Ohio | 1.7 | 16.5 |
| three | 2002 | Ohio | 3.6 | 16.5 |
| four | 2001 | Nevada | 2.4 | 16.5 |
| five | 2002 | Nevada | 2.9 | 16.5 |
| six | 2003 | Nevada | 3.2 | 16.5 |
1
2
| frame2['debt'] = np.arange(6.)
frame2
|
Output:
| | year | state | pop | debt |
| one | 2000 | Ohio | 1.5 | 0.0 |
| two | 2001 | Ohio | 1.7 | 1.0 |
| three | 2002 | Ohio | 3.6 | 2.0 |
| four | 2001 | Nevada | 2.4 | 3.0 |
| five | 2002 | Nevada | 2.9 | 4.0 |
| six | 2003 | Nevada | 3.2 | 5.0 |
Note: When assigning lists or arrays to a column, we must make sure that the value’s length must match the length of the DataFrame. If we assign a Series, its labels will be realigned exactly to the DataFrame’s index, inserting missing values in any holes
1
2
3
4
| val = pd.Series([-1.2,-1.5,-1.7], index = ['two', 'four', 'five'])
frame2['debt'] = val
frame2
|
Output:
| | year | state | pop | debt |
| one | 2000 | Ohio | 1.5 | NaN |
| two | 2001 | Ohio | 1.7 | -1.2 |
| three | 2002 | Ohio | 3.6 | NaN |
| four | 2001 | Nevada | 2.4 | -1.5 |
| five | 2002 | Nevada | 2.9 | -1.7 |
| six | 2003 | Nevada | 3.2 | NaN |
Assinging a column that doesn’t exist will create a new column. And the ‘del’ keyword wil delete columns as with a dict.
1
2
| frame2['eastern'] = (frame2.state == 'Ohio')
frame2
|
Output:
| | year | state | pop | debt | eastern |
| one | 2000 | Ohio | 1.5 | NaN | True |
| two | 2001 | Ohio | 1.7 | -1.2 | True |
| three | 2002 | Ohio | 3.6 | NaN | True |
| four | 2001 | Nevada | 2.4 | -1.5 | False |
| five | 2002 | Nevada | 2.9 | -1.7 | False |
| six | 2003 | Nevada | 3.2 | NaN | False |
1
2
| del frame2['eastern']
frame2
|
Output:
| | year | state | pop | debt |
| one | 2000 | Ohio | 1.5 | NaN |
| two | 2001 | Ohio | 1.7 | -1.2 |
| three | 2002 | Ohio | 3.6 | NaN |
| four | 2001 | Nevada | 2.4 | -1.5 |
| five | 2002 | Nevada | 2.9 | -1.7 |
| six | 2003 | Nevada | 3.2 | NaN |
1
2
| frame2.columns
Output: Index(['year', 'state', 'pop', 'debt'], dtype='object')
|
Note: The column returned from indexing a DataFrame is a view on the underlying data, not a copy. Thus, any in-place modifications to the Series will be reflected in the DataFrame. The column can be explicitely copied with the Series’s copy method.
1
2
| pop = {'Nevada': {2001: 2.4, 2002: 2.9},
'Ohio':{2000:1.5, 2001: 1.7, 2002: 3.6}}
|
If the nested dict is passed to the DataFrame, pandas will intrepret the outer dict keys as the columns and the inner dict keys as the rows indices
1
2
| frame3 = pd.DataFrame(pop)
frame3
|
Output:
| | Nevada | Ohio |
| 2001 | 2.4 | 1.7 |
| 2002 | 2.9 | 3.6 |
| 2000 | NaN | 1.5 |
We can transpose the DataFrame with similar syntax as in Numpy array. Here, the tranposed dataframe will just be a copy and not a view.
Output:
| | 2000 | 2001 | 2002 |
| Nevada | NaN | 2.4 | 2.9 |
| Ohio | 1.5 | 1.7 | 3.6 |
Output:
| | Nevada | Ohio |
| 2001 | 2.4 | 1.7 |
| 2002 | 2.9 | 3.6 |
| 2000 | NaN | 1.5 |
The keys in the inner dicts are combined and sorted to form the index in the result. This isn’t true if an explicit index is specified:
1
| pd.DataFrame(pop, index = [2001,2002,2003])
|
Output:
| | Nevada | Ohio |
| 2001 | 2.4 | 1.7 |
| 2002 | 2.9 | 3.6 |
| 2003 | NaN | NaN |
The Dict of Sereis are treated in much the same way
1
2
3
| pdata = {'Ohio': frame3['Ohio'][:-1],
'Nevada':frame3['Nevada'][:2]}
pd.DataFrame(pdata)
|
Output:
| | Nevada | Ohio |
| 2000 | NaN | 1.5 |
| 2001 | 2.4 | 1.7 |
Output:
| | Nevada | Ohio |
| 2001 | 2.4 | 1.7 |
| 2002 | 2.9 | 3.6 |
| 2000 | NaN | 1.5 |
If a DataFrame’s index and columns have their name attribtues set, these will also be displayed along with the frame
1
2
| frame3.index.name = 'year'; frame3.columns.name = 'state'
frame3
|
Output:
| state | Nevada | Ohio |
| year | | |
| 2000 | NaN | 1.5 |
| 2001 | 2.4 | 1.7 |
| 2002 | 2.9 | 3.6 |
As with Series, the values attribute returns the data contained in the DataFrame as a two-dimensional ndarray
1
2
3
4
5
| frame3.values
Output:
array([[2.4, 1.7],
[2.9, 3.6],
[nan, 1.5]])
|
1
2
3
4
5
6
7
8
| frame2.values
Output:
array([[2000, 'Ohio', 1.5, nan],
[2001, 'Ohio', 1.7, -1.2],
[2002, 'Ohio', 3.6, nan],
[2001, 'Nevada', 2.4, -1.5],
[2002, 'Nevada', 2.9, -1.7],
[2003, 'Nevada', 3.2, nan]], dtype=object)
|
![img]()
Index Objects
Pandas’s Index objects are responsible for holding the axis lables and other metadata. Any array or other sequence of labels we use when constructing a Series or DataFrame is internally convertedto an Index:
1
2
3
4
5
6
7
| obj = pd.Series(range(3), index = ['a','b', 'c'])
index = obj.index
index
Output: Index(['a', 'b', 'c'], dtype='object')
index[1:]
Output: Index(['b', 'c'], dtype='object')
|
Index objects are IMMUTABLE and thus can’t be modified by the user. Thus makes it safer to share Index objects among data structures.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
| labels = pd.Index(np.arange(3))
labels
Output: Int64Index([0, 1, 2], dtype='int64')
obj2 = pd.Series([1.5, 2.5, 0],index = labels)
obj2
Output:
0 1.5
1 2.5
2 0.0
dtype: float64
obj2.index is labels
Output: True
|
Some users will not often take advantage of the capabilities pro‐ vided by indexes, but because some operations will yield results containing indexed data, it’s important to understand how they work.
Output:
| state | Nevada | Ohio |
| year | | |
| 2001 | 2.4 | 1.7 |
| 2002 | 2.9 | 3.6 |
| 2000 | NaN | 1.5 |
Note: In addition to being array-like, an Index also behaves like a fixed size set
1
2
| frame3.columns
Output: Index(['Nevada', 'Ohio'], dtype='object', name='state')
|
1
2
3
4
5
| 'Ohio' in frame3.columns
Output: True
2003 in frame3.index
Output: False
|
Note: Unlike python sets, a pandas Index can contain duplicate labels
1
2
3
4
| dup_labels = pd.Index(['foo', 'foo', 'bar', 'bar'])
dup_labels
Output: Index(['foo', 'foo', 'bar', 'bar'], dtype='object')
|
But selections with duplicate labels will select all occurences of that label.
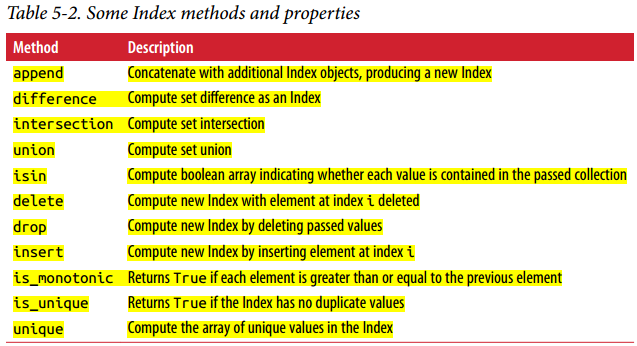
Each Index has a numer of methods and properteis for set logic, which answer other common questions about the data it contains.
![img]()
Essential Functionality
Reindexing - An important method on pandas objects is reindex, which means to create a new object with the data conformed to a new index.
1
2
3
4
5
6
7
8
9
| obj = pd.Series([4.5, 7.2, -5.3, 3.6], index = ['d', 'b', 'a', 'c'])
obj
Output:
d 4.5
b 7.2
a -5.3
c 3.6
dtype: float64
|
Calling reindex on the above Series rearranges the data according to the new index, introducing missing values if any index values were not already present.
1
2
3
4
5
6
7
8
9
| obj2 = obj.reindex(['a', 'b', 'c', 'd', 'e'])
obj2
Output:
a -5.3
b 7.2
c 3.6
d 4.5
e NaN
dtype: float64
|
Note: For ordered data like time series, it may be desirable to do some interpolation or filling of values when reindexing. The method option allows us to do this, using a method such as ffill, which forward-fills the values:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
| obj3 = pd.Series(['blue', 'purple', 'yellow'], index = [0,2,4])
Output:
0 blue
2 purple
4 yellow
dtype: object
obj3.reindex(range(6), method = 'ffill')
Output:
0 blue
1 blue
2 purple
3 purple
4 yellow
5 yellow
dtype: object
|
Note: With DataFrame, reindex can alter either the (row) index, columns or both. When passed only a sequence, it reindexed the rows in the result:
1
2
3
4
| frame = pd.DataFrame(np.arange(9).reshape((3,3)),
index = ['a', 'c', 'd'],
columns = ['Ohio', 'Texas', 'California'])
frame
|
Output:
| | Ohio | Texas | California |
| a | 0 | 1 | 2 |
| c | 3 | 4 | 5 |
| d | 6 | 7 | 8 |
1
2
| frame2 = frame.reindex(['a', 'b', 'c', 'd'])
frame2
|
Output:
| | Ohio | Texas | California |
| a | 0 | 1 | 2 |
| b | NaN | NaN | NaN |
| c | 3 | 4 | 5 |
| d | 6 | 7 | 8 |
The columns can be reindexed with the columns keyword
1
2
3
| states = ['Texas', 'Utah', 'California']
frame.reindex(columns=states)
|
Output:
| | Texas | Utah | California |
| a | 1 | NaN | 2 |
| c | 4 | NaN | 5 |
| d | 7 | NaN | 8 |
Note: We can reindex more succinctly by lable-indexing withloc, and this way is more preferable by many users.
![img]()
Dropping Entries from an Axis
Dropping one or more entries from an axis is easy if you already have an index array or list without those entries. As that can require a bit of munging and set logic, the drop method will return a new object with the indicated value or values deleted from an axis:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
| obj = pd.Series(np.arange(5.), index = ['a', 'b', 'c','d', 'e'])
obj
Output:
a 0.0
b 1.0
c 2.0
d 3.0
e 4.0
dtype: float64
new_obj = obj.drop('c')
new_obj
Output:
a 0.0
b 1.0
d 3.0
e 4.0
dtype: float64
obj.drop(['d', 'c'])
Output:
a 0.0
b 1.0
e 4.0
dtype: float64
|
With DataFrame, index values can be deleted from either axis.
1
2
3
4
| data = pd.DataFrame(np.arange(16).reshape((4,4)),
index = ['Ohio', 'Colorado', 'Utah', 'New York'],
columns = ['one', 'two', 'three', 'four'])
data
|
Output:
| | one | two | three | four |
| Ohio | 0 | 1 | 2 | 3 |
| Colorado | 4 | 5 | 6 | 7 |
| Utah | 8 | 9 | 10 | 11 |
| New York | 12 | 13 | 14 | 15 |
Calling drop with a sequence of lables will drop values from the row labels(axis = 0)
1
| data.drop(['Colorado', 'Ohio'])
|
Output:
| | one | two | three | four |
| Utah | 8 | 9 | 10 | 11 |
| New York | 12 | 13 | 14 | 15 |
We can drop values from the columns by passing axis = 1 or axis = 'columns'
1
| data.drop('two', axis = 1)
|
Output:
| | one | three | four |
| Ohio | 0 | 2 | 3 |
| Colorado | 4 | 6 | 7 |
| Utah | 8 | 10 | 11 |
| New York | 12 | 14 | 15 |
1
| data.drop('three', axis = 'columns')
|
Output:
| | one | two | four |
| Ohio | 0 | 1 | 3 |
| Colorado | 4 | 5 | 7 |
| Utah | 8 | 9 | 11 |
| New York | 12 | 13 | 15 |
Many functions like drop, which modify the size or shape of a Series or DataFrame, can manipulate an object in-place without returning a new object.
1
2
3
4
5
6
7
8
9
| obj.drop('c', inplace=True)
obj
Output:
a 0.0
b 1.0
d 3.0
e 4.0
dtype: float64
|
Note: Be careful with the inplace, as it destroys any data that is dropped.
Indexing, Selection, and Filtering
Series indexing (obj[…]) works analogously to NumPy array indexing, except you can use the Series’s index values instead of only integers.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
| obj = pd.Series(np.arange(4.), index=['a','b', 'c','d'])
obj
Output:
a 0.0
b 1.0
c 2.0
d 3.0
dtype: float64
obj['b']
Output: 1.0
obj[2]
Output: 2.0
obj[2:4]
Output:
c 2.0
d 3.0
dtype: float64
obj[['b', 'a', 'd']]
Output:
b 1.0
a 0.0
d 3.0
dtype: float64
obj[[1,3]]
Output:
b 1.0
d 3.0
dtype: float64
obj[obj<2]
Output:
a 0.0
b 1.0
dtype: float64
|
Slicing with lables behaves differently than normal Python slicing in that the end-points are inclusive.
1
2
3
4
5
| obj['b' : 'c']
Output:
b 1.0
c 2.0
dtype: float64
|
Setting using these methods modifies the corresponding section of the Series
1
2
3
4
5
6
7
8
| obj['b':'c'] = 5
obj
Output:
a 0.0
b 5.0
c 5.0
d 3.0
dtype: float64
|
Indexing into a DataFrame is for retrieving one or more columns either with a single vlaue or sequence
1
2
3
4
| data = pd.DataFrame(np.arange(16).reshape((4,4)),
index = ['Ohio', 'Colorado', 'Utah', 'New York'],
columns = ['one', 'two', 'three', 'four'])
data
|
Output:
| | one | two | three | four |
| Ohio | 0 | 1 | 2 | 3 |
| Colorado | 4 | 5 | 6 | 7 |
| Utah | 8 | 9 | 10 | 11 |
| New York | 12 | 13 | 14 | 15 |
1
2
3
4
5
6
7
| data['two']
Output:
Ohio 1
Colorado 5
Utah 9
New York 13
Name: two, dtype: int32
|
1
2
3
4
5
6
7
8
| data [['three', 'one']]
Output:
three one
Ohio 1 0
Colorado 5 4
Utah 9 8
New York 13 12
Name: two, dtype: int32
|
Indexing like this has a few special cases. First, slicing or selecting data with a boolean array:
Output:
| | one | two | three | four |
| Ohio | 0 | 1 | 2 | 3 |
| Colorado | 4 | 5 | 6 | 7 |
Output:
| | one | two | three | four |
| Ohio | 0 | 1 | 2 | 3 |
| Colorado | 4 | 5 | 6 | 7 |
| New York | 12 | 13 | 14 | 15 |
The row selection syntax data[:2] is provided as a convenience. Passing a single element or a list to the [] operator selects columns.
Another use case is in indexing with a boolean DataFrame, such as one produced by a scalar comparision
Output:
| | one | two | three | four |
| Ohio | 0 | 1 | 2 | 3 |
| Colorado | 4 | 5 | 6 | 7 |
| Utah | 8 | 9 | 10 | 11 |
| New York | 12 | 13 | 14 | 15 |
Output:
| | one | two | three | four |
| Ohio | True | True | True | True |
| Colorado | True | False | False | False |
| Utah | False | False | False | False |
| New York | False | False | False | False |
1
2
| data[data<5] = 0
data
|
Output:
| | one | two | three | four |
| Ohio | 0 | 0 | 0 | 0 |
| Colorado | 0 | 5 | 6 | 7 |
| Utah | 8 | 9 | 10 | 11 |
| New York | 12 | 13 | 14 | 15 |
This makes DataFrame syntactically more like a two-dimensional NumPy array in this particular case.
Selection with loc and iloc
Output:
| | one | two | three | four |
| Ohio | 0 | 0 | 0 | 0 |
| Colorado | 0 | 5 | 6 | 7 |
| Utah | 8 | 9 | 10 | 11 |
| New York | 12 | 13 | 14 | 15 |
The special indexing operators loc and iloc enables us to select a subset of rows and columns from a DataFrame with NumPy-like notation using either axis labels(loc) or integers (iloc)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
| data.loc['Colorado', ['two', 'three']]
Output:
two 5
three 6
Name: Colorado, dtype: int32
data.iloc[2,[3,0,1]]
Output:
four 11
one 8
two 9
Name: Utah, dtype: int32
data.iloc[2]
Output:
one 8
two 9
three 10
four 11
Name: Utah, dtype: int32
|
1
| data.iloc[[1,2],[3,0,1]]
|
Output:
| | four | one | tow |
| Colorado | 7 | 0 | 5 |
| Utah | 11 | 8 | 9 |
Both indexing functions work with slices in addition to single labels or list of labels
Output:
| | one | two | three | four |
| Ohio | 0 | 0 | 0 | 0 |
| Colorado | 0 | 5 | 6 | 7 |
| Utah | 8 | 9 | 10 | 11 |
| New York | 12 | 13 | 14 | 15 |
1
2
3
4
5
6
| data.loc[:'Utah', 'two']
Output:
Ohio 0
Colorado 5
Utah 9
Name: two, dtype: int32
|
1
| data.iloc[:, :3][data['three'] > 5]
|
Output:
| | one | two | three |
| Colorado | 0 | 5 | 6 |
| Utah | 8 | 9 | 10 |
| New York | 12 | 13 | 14 |
![img]()
![img]()
Integer Indexes
1
2
3
4
5
6
7
| ser = pd.Series(np.arange(3.))
ser
Output:
0 0.0
1 1.0
2 2.0
dtype: float64
|
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
| ser2 = pd.Series(np.arange(3.), index = ['a', 'b', 'c'])
ser2
Output:
a 0.0
b 1.0
c 2.0
dtype: float64
ser2[-1]
Output: 2.0
ser[:1]
Output:
0 0.0
dtype: float64
ser.loc[:1]
Output:
0 0.0
1 1.0
dtype: float64
ser.iloc[:1]
Output:
0 0.0
dtype: float64
|
Arithmetic and Data Alignment
An important pandas feature for some applications is the behavior of arithmetic between objects with different indexes. When you are adding together objects, if any index pairs are not the same, the respective index in the result will be the union of the index paris. For users with database experience, this is similar to an automatic Outer Join on the index labels.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
| s1 = pd.Series([7.3, -2.5, 3.4, 1.5], index = ['a', 'c', 'd', 'e'])
s2 = pd.Series([-2.1, 3.6, -1.5, 4, 3.1], index = ['a', 'c', 'e', 'f', 'g'])
s1
Output:
a 7.3
c -2.5
d 3.4
e 1.5
dtype: float64
s2
Output:
a -2.1
c 3.6
e -1.5
f 4.0
g 3.1
dtype: float64
s1 + s2
Output:
a 5.2
c 1.1
d NaN
e 0.0
f NaN
g NaN
dtype: float64
|
Here, the internal data alignment introduces missing values in the label locations that don’t overlap. Missing values will then propagate in further arithmetic computations.
In case of DataFrame, alignment is performed on both the rows and the columns
1
2
3
4
5
6
7
8
9
| df1 = pd.DataFrame(np.arange(9.).reshape((3,3)),
columns = list('bcd'),
index = ['Ohio', 'Texas', 'Colorado'])
df2 = pd.DataFrame(np.arange(12.).reshape((4,3)),
columns = list('bde'),
index = ['Utah', 'Ohio', 'Texas', 'Oregon'])
df1
|
Output:
| | b | c | d |
| Ohio | 0.0 | 1.0 | 2.0 |
| Texas | 3.0 | 4.0 | 5.0 |
| Colorado | 6.0 | 7.0 | 8.0 |
Output:
| | b | d | e |
| Ohio | 0.0 | 1.0 | 2.0 |
| Texas | 3.0 | 4.0 | 5.0 |
| Colorado | 6.0 | 7.0 | 8.0 |
| Oregon | 9.0 | 10.0 | 11.0 |
Adding these together returns a DataFrame whose index and columns are the unionis of the ones in each DataFrame
Output:
| | b | c | d | e |
| Colorado | NaN | NaN | NaN | NaN |
| Ohio | 3.0 | NaN | 6.0 | NaN |
| Oregon | NaN | NaN | NaN | NaN |
| Texas | 9.0 | NaN | 12.0 | NaN |
| Utah | NaN | NaN | NaN | NaN |
Since, the ‘c’ and ‘e’ columns are note present in bothe the DataFrame objects, they appears as all missing in the result. The same holds for the ‘Utah’, ‘Colorado’, and ‘Oregon’ whose labels are not common to both objects.
Note: If you add DataFrame objects with no column or row labels in common, then the result will contain all nulls:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
| df1 = pd.DataFrame({'A': [1,2]})
df2 = pd.DataFrame({'B' : [3,4]})
df1
Output:
A
0 1
1 2
df2
Output:
B
0 3
1 4
df1 - df2
A B
0 NaN NaN
1 NaN NaN
|
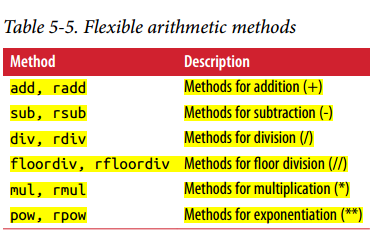
Arithmetic methods with fill values
In arithmetic operations between differently indexed objects, you might want to fill with a special value, like 0, when an axis lable is found in one object but not the other
1
2
3
4
5
6
| df1 = pd.DataFrame(np.arange(12.).reshape((3,4)),
columns = list('abcd'))
df2 = pd.DataFrame(np.arange(20.).reshape((4,5)),
columns=list('abcde'))
df2.loc[1, 'b'] = np.nan
|
Output:
| | a | b | c | d |
| 0 | 0.0 | 1.0 | 2.0 | 3.0 |
| 1 | 4.0 | 5.0 | 6.0 | 7.0 |
| 2 | 8.0 | 9.0 | 10.0 | 11.0 |
Output:
| | a | b | c | d | e |
| 0 | 0.0 | 1.0 | 2.0 | 3.0 | 4.0 |
| 1 | 5.0 | NaN | 7.0 | 8.0 | 9.0 |
| 2 | 10.0 | 11.0 | 12.0 | 13.0 | 14.0 |
| 3 | 15.0 | 16.0 | 17.0 | 18.0 | 19.0 |
Adding these together results in NA values in the locations that don’t overlap:
Output:
| | a | b | c | d | e |
| 0 | 0.0 | 2.0 | 4.0 | 6.0 | NaN |
| 1 | 9.0 | NaN | 13.0 | 15.0 | NaN |
| 2 | 18.0 | 20.0 | 22.0 | 24.0 | NaN |
| 3 | NaN | NaN | NaN | NaN | NaN |
Using the add method on df1,we can pass df2 and an arguemnt to fill_value
1
| df1.add(df2, fill_value = 0)
|
Output:
| | a | b | c | d | e |
| 0 | 0.0 | 2.0 | 4.0 | 6.0 | 4.0 |
| 1 | 9.0 | 5..0 | 13.0 | 15.0 | 9.0 |
| 2 | 18.0 | 20.0 | 22.0 | 24.0 | 14.0 |
| 3 | 15.0 | 16.0 | 17.0 | 18.0 | 19.0 |
Output:
| | a | b | c | d |
| 0 | 0.0 | 1.0 | 2.0 | 3.0 |
| 1 | 4.0 | 5.0 | 6.0 | 7.0 |
| 2 | 8.0 | 9.0 | 10.0 | 11.0 |
Output:
| | a | b | c | d |
| 0 | inf | 1.000000 | 0.500000 | 0.333333 |
| 1 | 0.250000 | 0.200000 | 0.166667 | 0.142857 |
| 2 | 0.125000 | 0.111111 | 0.100000 | 0.090909 |
Output:
| | a | b | c | d |
| 0 | inf | 1.000000 | 0.500000 | 0.333333 |
| 1 | 0.250000 | 0.200000 | 0.166667 | 0.142857 |
| 2 | 0.125000 | 0.111111 | 0.100000 | 0.090909 |
When reindxing a Series or DataFrame, we can speciy a different fill value
1
| df1.reindex(columns = df2.columns, fill_value=0)
|
Output:
| | a | b | c | d | e |
| 0 | 0.0 | 1.0 | 2.0 | 3.0 | 0 |
| 1 | 4.0 | 5.0 | 6.0 | 7.0 | 0 |
| 2 | 8.0 | 9.0 | 10.0 | 11.0 | 0 |
![img]()
Operations between DataFrame and Series
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
| arr = np.arange(12.).reshape((3,4))
arr
Output:
array([[ 0., 1., 2., 3.],
[ 4., 5., 6., 7.],
[ 8., 9., 10., 11.]])
arr[0]
Output: array([0., 1., 2., 3.])
arr - arr[0]
Output:
array([[0., 0., 0., 0.],
[4., 4., 4., 4.],
[8., 8., 8., 8.]])
|
Here, when we subtract arr[0] from arr, the subtraction is performed once for each row. This is referred to as broadcasting.
1
2
3
4
5
| frame = pd.DataFrame(np.arange(12.).reshape((4,3)),
columns = list('bde'),
index = ['Utah', 'Ohio', 'Teas', 'Oregon'])
series = frame.iloc[0]
frame
|
Output:
| | b | d | e |
| Utah | 0.0 | 1.0 | 2.0 |
| Ohio | 3.0 | 4.0 | 5.0 |
| Texas | 6.0 | 7.0 | 8.0 |
| Oregon | 9.0 | 10.0 | 11.0 |
1
2
3
4
5
6
| series
Output:
b 0.0
d 1.0
e 2.0
Name: Utah, dtype: float64
|
By default, arithmetic between a DataFrame and Series matches the index of the Series on the DataFrame’s columns, broadcasting down the rows
Output:
| | b | d | e |
| Utah | 0.0 | 0.0 | 0.0 |
| Ohio | 3.0 | 3.0 | 3.0 |
| Texas | 6.0 | 6.0 | 6.0 |
| Oregon | 9.0 | 9.0 | 9.0 |
If an index value is not found in either the DataFrame’s columns or the Series’s index, then the objects will be reindexed to form the union
1
2
| series2 = pd.Series(range(3), index = ['b', 'e', 'f'])
frame + series2
|
Output:
| | b | d | e | f |
| Utah | 0.0 | NaN | 3.0 | NaN |
| Ohio | 3.0 | NaN | 6.0 | NaN |
| Texas | 6.0 | NaN | 9.0 | NaN |
| Oregon | 9.0 | NaN | 12.0 | NaN |
If we want to instead broadcast over the columns, matching on the rows, we have to use one of the arithmetic methods
Output:
| | b | d | e |
| Utah | 0.0 | 1.0 | 2.0 |
| Ohio | 3.0 | 4.0 | 5.0 |
| Texas | 6.0 | 7.0 | 8.0 |
| Oregon | 9.0 | 10.0 | 11.0 |
1
2
3
4
5
6
7
8
| series3 = frame['d']
series3
Output:
Utah 1.0
Ohio 4.0
Teas 7.0
Oregon 10.0
Name: d, dtype: float64
|
1
| frame.sub(series3, axis = 'index')
|
Output:
| | b | d | e |
| Utah | -1.0 | 0.0 | 1.0 |
| Ohio | -1.0 | 0.0 | 1.0 |
| Texas | -1.0 | 0.0 | 1.0 |
| Oregon | -1.0 | 0.0 | 1.0 |
The axis number that we pass is the axis to mathc on. In this case we mean to match on the DataFrame’s row index (axis=’index’ or axis= 0) and broadcast across the columns.
Function Application and Mapping
Numpy ufuncs (element-wise array methods) also work with pandas objects
1
2
3
4
5
6
7
8
9
10
11
| frame = pd.DataFrame(np.random.randn(4,3),
columns = list('bde'),
index = ['Utah', 'Ohio', 'Texas', 'Oregon'])
frame
Output:
b d e
Utah -0.059728 -1.671352 -2.322987
Ohio -1.072084 -1.265158 -1.452127
Texas -1.487410 -2.289852 -1.427222
Oregon -0.852068 -0.911926 0.486711
|
1
2
3
4
5
6
7
| np.abs(frame)
Output:
b d e
Utah 0.059728 1.671352 2.322987
Ohio 1.072084 1.265158 1.452127
Texas 1.487410 2.289852 1.427222
Oregon 0.852068 0.911926 0.486711
|
Another frequent operation is applying a function on one-dimensional arrays to each column or row. DataFrame’s apply method does exactly this.
1
2
3
4
5
6
7
| f = lambda x: x.max() - x.min()
frame.apply(f)
Output:
b 1.427681
d 1.377926
e 2.809697
dtype: float64
|
Here, the function f, which computes the difference between the maximum and minimum of a Series, is invoked once on each column in frame. The result is a Series having the columns of frame as its index.
If we pass axis = 'columns' to apply, the function will be invoked once per row instead
1
2
3
4
5
6
7
| frame.apply(f, axis = 'columns')
Output:
Utah 2.263259
Ohio 0.380044
Texas 0.862630
Oregon 1.398637
dtype: float64
|
Many of the most common array statistics (like sum and mean) are DataFrame methods so using apply is not necessary.
The function passed to apply need not return a scalar value, it can also return a Series with multiple values. For example:
1
2
3
4
5
6
7
8
9
| def f(x):
return pd.Series([x.min(),x.max()],
index = ['min', 'max'])
frame.apply(f)
Output:
b d e
min -1.487410 -2.289852 -2.322987
max -0.059728 -0.911926 0.486711
|
1
2
3
4
5
6
7
8
| frame.apply(f, axis = 'columns')
Output:
min max
Utah -2.322987 -0.059728
Ohio -1.452127 -1.072084
Texas -2.289852 -1.427222
Oregon -0.911926 0.486711
|
Element wise python functions can be used, too. Suppose, you wanted to compute a formatted string from each floating-point value in frame. You can do this with applymap
1
2
3
4
5
6
7
8
9
| format = lambda x: '%2f' % x
frame.applymap(format)
Output:
b d e
Utah -0.059728 -1.671352 -2.322987
Ohio -1.072084 -1.265158 -1.452127
Texas -1.487410 -2.289852 -1.427222
Oregon -0.852068 -0.911926 0.486711
|
The reason for the name applymap is that Series has a map method for applying an element-wise function
1
2
3
4
5
6
7
| frame['e'].map(format)
Output:
Utah -2.322987
Ohio -1.452127
Texas -1.427222
Oregon 0.486711
Name: e, dtype: object
|
Sorting and Ranking
Sorting a dataset by some criterion is another important built-in operation. To sort lexicographically by row or column index, use the sort_index method, which returns a new, sorted object
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
| obj = pd.Series(range(4), index = ['d', 'a','b','c'])
obj
Output:
d 0
a 1
b 2
c 3
dtype: int64
obj.sort_index()
Output:
a 1
b 2
c 3
d 0
dtype: int64
|
With a DataFrame, we can sort by index on either axis
1
2
3
4
5
6
7
8
9
| frame = pd.DataFrame(np.arange(8).reshape((2,4)),
index = ['three', 'one'],
columns = ['d', 'a', 'b', 'c'])
frame
Output:
d a b c
three 0 1 2 3
one 4 5 6 7
|
1
2
3
4
5
6
| frame.sort_index()
Output:
d a b c
one 4 5 6 7
three 0 1 2 3
|
1
2
3
4
5
6
| frame.sort_index(axis = 1)
Output:
a b c d
three 1 2 3 0
one 5 6 7 4
|
The data is sorted in ascending order by default, but can be sorted in descending order too using the ascending attribute
1
2
3
4
5
6
| frame.sort_index(axis = 1, ascending=False)
Output:
d c b a
three 0 3 2 1
one 4 7 6 5
|
To sort a Series by its values, use it’s sort_values method
1
2
3
4
5
6
7
8
| obj = pd.Series([4,7,-3,2])
obj.sort_values()
Output:
2 -3
3 2
0 4
1 7
dtype: int64
|
Any missing values are sorted to the end of the Series by default
1
2
3
4
5
6
7
8
9
10
| obj = pd.Series([4, np.nan, 7, np.nan, -3, 2])
obj.sort_values()
Output:
4 -3.0
5 2.0
0 4.0
2 7.0
1 NaN
3 NaN
dtype: float64
|
When sorting a DataFrame, we can use the data in one or more columns as the sort keys.To do so, pass one or more columns names to the by option of sort_values
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
| frame = pd.DataFrame({'b': [4,7,-3,2,], 'a': [0,1,0,1]})
frame
Output:
b a
0 4 0
1 7 1
2 -3 0
3 2 1
frame.sort_values(by='a')
Output:
b a
0 4 0
2 -3 0
1 7 1
3 2 1
|
To sort by multiple columns, pass a list of name
1
2
3
4
5
6
7
8
| frame.sort_values(by=['a', 'b'])
Output:
b a
2 -3 0
0 4 0
3 2 1
1 7 1
|
Ranking assigns ranks from one through the number of valid data points in an array. The rank methods for Series and DataFrame are the place to look; by default rank breaks ties by assinging each group the mean rank
1
2
3
4
5
6
7
8
9
10
11
| obj = pd.Series([7,-5,7,4,2,0,4])
obj.rank()
Output:
0 6.5
1 1.0
2 6.5
3 4.5
4 3.0
5 2.0
6 4.5
dtype: float64
|
Ranks can also be assigned according to the order in which they’re observed in the data
1
2
3
4
5
6
7
8
9
10
| obj.rank(method='first')
Output:
0 6.0
1 1.0
2 7.0
3 4.0
4 3.0
5 2.0
6 5.0
dtype: float64
|
Here, instead of using the average rank 6.5 for the entries 0 and 2, they instead have been set ot 6 and 7 because label 0 precedes label 2 in the data.
We can rank in descending order too
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
| obj
Output:
0 7
1 -5
2 7
3 4
4 2
5 0
6 4
dtype: int64
obj.rank(ascending=False, method = 'max')
Output:
0 2.0
1 7.0
2 2.0
3 4.0
4 5.0
5 6.0
6 4.0
dtype: float64
|
DataFrame can compute ranks over the rows or the columns
1
2
3
4
5
6
7
8
9
10
| frame = pd.DataFrame({'b': [4.3, 7, -3,2], 'a': [0,1,0,1],
'c': [-2,5,8,-2.5]})
frame
Output:
b a c
0 4.3 0 -2.0
1 7.0 1 5.0
2 -3.0 0 8.0
3 2.0 1 -2.5
|
1
2
3
4
5
6
7
8
| frame.rank(axis='columns')
Output:
b a c
0 3.0 2.0 1.0
1 3.0 1.0 2.0
2 1.0 2.0 3.0
3 3.0 2.0 1.0
|
![img]()
Axis Indexes with Duplicate Labels
1
2
3
4
5
6
7
8
9
| obj = pd.Series(range(5), index = ['a', 'a', 'b','b', 'c'])
obj
Output:
a 0
a 1
b 2
b 3
c 4
dtype: int64
|
The index’s is_unique property can tell you whether its labels are unique or not
1
2
| obj.index.is_unique
Output: False
|
Data Selection is one of the main things that behaves differently with duplicates. Indexing a label with multiple entries returns a Series, while single entries return a scalar value.
1
2
3
4
5
6
7
8
| obj['a']
Output:
a 0
a 1
dtype: int64
obj['c']
Output: 4
|
This can make our code more complicated as the output type from indexing can vary based on whether a label is repeated or not.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
| df = pd.DataFrame(np.random.randn(4,3),
index = ['a', 'a', 'b', 'b'])
df
Output:
0 1 2
a -0.088814 -0.602398 0.402683
a 1.195694 -0.383322 0.330696
b -3.542210 0.460190 0.339993
b -0.718968 -0.049578 0.127387
df.index.is_unique
Output: False
df.loc['b']
Output:
0 1 2
b -3.542210 0.460190 0.339993
b -0.718968 -0.049578 0.127387
|
Summarizing and Computing Descriptive Statistics
pandas objects are equipped with a set of common mathematical and statistical meth‐ ods. Most of these fall into the category of reductions or summary statistics, methods that extract a single value (like the sum or mean) from a Series or a Series of values from the rows or columns of a DataFrame.
1
2
3
4
5
6
7
8
9
10
11
12
| df = pd.DataFrame([[1.4, np.nan], [7.1, -4.5],
[np.nan, np.nan], [0.75, -1.3]],
index = ['a', 'b', 'c', 'd'],
columns = ['one', 'two'])
df
Output:
one two
a 1.40 NaN
b 7.10 -4.5
c NaN NaN
d 0.75 -1.3
|
Calling DataFrame’s sum method returns a Series containing Column Sums
1
2
3
4
5
| df.sum()
Output:
one 9.25
two -5.80
dtype: float64
|
Passing axis = 'columns' or axis = 1 sums across the columsn instead
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
| df.sum(axis='columns')
Output:
a 1.40
b 2.60
c 0.00
d -0.55
dtype: float64
df.sum(axis='columns',skipna=False)
Output:
a NaN
b 2.60
c NaN
d -0.55
dtype: float64
|
NA values are excluded unless the entire slice (row or column in this case) is NA. This can be disabled with the skipna option
1
2
3
4
5
6
7
| df.mean(axis='columns', skipna= False)
Output:
a NaN
b 1.300
c NaN
d -0.275
dtype: float64
|
![img]()
Some methods like idxmin and idxmax return indirect statistics like the index value where the minimum or maximum values are attained:
1
2
3
4
5
| df.idxmax()
Output:
one b
two d
dtype: object
|
Other methods are accumulations
1
2
3
4
5
6
7
8
| df.cumsum()
Output:
one two
a 1.40 NaN
b 8.50 -4.5
c NaN NaN
d 9.25 -5.8
|
Another type of method is neither a reduction nor an accumulatoin. describeis one such example, producing multiple summary statistics in one shot.
1
2
3
4
5
6
7
8
9
10
11
12
| df.describe()
Output:
one two
count 3.000000 2.000000
mean 3.083333 -2.900000
std 3.493685 2.262742
min 0.750000 -4.500000
25% 1.075000 -3.700000
50% 1.400000 -2.900000
75% 4.250000 -2.100000
max 7.100000 -1.300000
|
On non-numeric data, describe produces alternative summary statistics
1
| obj = pd.Series(['a', 'a', 'b', 'c'] * 4)
|
1
2
3
4
5
6
7
| obj.describe()
Output:
count 16
unique 3
top a
freq 8
dtype: object
|
![img]()
Unique Values, Value Counts, and Membership
1
2
3
4
| obj = pd.Series(['c', 'a', 'd', 'a', 'a', 'b', 'b', 'c', 'c'])
uniques = obj.unique()
uniques
Output: array(['c', 'a', 'd', 'b'], dtype=object)
|
The unique values are not necessarily returned in sorted order, but could be sorted after the fact if needed (uniques.sort()). Relatedly, value_counts computes a Series containing value frequencies
1
2
3
4
5
6
7
| obj.value_counts()
Output:
a 3
c 3
b 2
d 1
dtype: int64
|
The Series is sorted by value in descending order as a convenience. But it can be sorted in ascending order as well by setting the attribute of sort value to False
1
2
3
4
5
6
7
| pd.value_counts(obj.values, sort = False)
Output:
a 3
d 1
c 3
b 2
dtype: int64
|
isin performs a vectorized set memebership check and can be useful in filtering a dataset down to a subset of values in a Series or column in a Dataframe
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
| obj
Output:
0 c
1 a
2 d
3 a
4 a
5 b
6 b
7 c
8 c
dtype: object
mask = obj.isin(['b', 'c'])
mask
Output:
0 True
1 False
2 False
3 False
4 False
5 True
6 True
7 True
8 True
dtype: bool
|
1
2
3
4
| #equivalent numpy expression
np.in1d(obj,['b','c'])
Output
array([ True, False, False, False, False, True, True, True, True])
|
1
2
3
4
5
6
7
8
| obj[mask]
Output:
0 c
5 b
6 b
7 c
8 c
dtype: object
|
Related to isin is the Index.get_indexer method, which gives us an index array from an array of possibly non-distinct values into another array of distinct values
1
2
3
4
| to_match = pd.Series(['c', 'a', 'b', 'b', 'c', 'a'])
unique_vals = pd.Series(['c', 'b', 'a'])
pd.Index(unique_vals).get_indexer(to_match)
Output: array([0, 2, 1, 1, 0, 2], dtype=int64)
|
![img]()
In some cases, we may want to compute a histogram on multiple related columns in a DataFrame.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
| data = pd.DataFrame({'Qu1': [1,3,4,3,4],
'Qu2': [2,3,1,2,3],
'Qu3': [1,5,2,4,4]})
data
Output:
Qu1 Qu2 Qu3
0 1 2 1
1 3 3 5
2 4 1 2
3 3 2 4
4 4 3 4
data.apply(pd.value_counts)
Output:
Qu1 Qu2 Qu3
1 1.0 1.0 1.0
2 NaN 2.0 1.0
3 2.0 2.0 NaN
4 2.0 NaN 2.0
5 NaN NaN 1.0
result = data.apply(pd.value_counts).fillna(0)
result
Output:
Qu1 Qu2 Qu3
1 1.0 1.0 1.0
2 0.0 2.0 1.0
3 2.0 2.0 0.0
4 2.0 0.0 2.0
5 0.0 0.0 1.0
|